Finding more information about a gene (September 6, 2005).
This page is moving to a new website.
I ran a few simple experiments using microarray data from a public source:
This is the data set used in the publication
- Database of mRNA gene expression profiles of multiple human organs. Son CG, Bilke S, Davis S, Greer BT, Wei JS, Whiteford CC, Chen QR, Cenacchi N, Khan J. Genome Res 2005: 15(3); 443-50. [Medline] [Abstract] [Full text] [PDF]
This data set is:
a high-density gene expression database of 18,927 unique genes for 158 normal human samples from 19 different organs of 30 different individuals using DNA microarrays.
I wanted to run a simple analysis to see what I could discover. I selected the 5 ovary tissue samples and compared them to the 7 testes tissue samples. Then, I analyzed the data and selected the 50 genes with the greatest evidence of differential expression. These 50 genes had the following CloneID values:
462333 2476256 190732 1642722 470227 2911881 345656 1751590 2092597 264868 726661 1550909 884658 276712 1751408 1550909 67037 757222 291290 1625698 377799 260170 1461577 795739 1469872 969769 436765 824479 1751408 1733262 38465 782306 197414 782141 726628 1502819 1468310 1469407 1049033 345342 1417595 1628729 781510 275771 782756 812161 345342 162491 755881 110307The question then becomes: what can I infer from this list? It is just a series of cryptic numbers. This is a difficult question for me to answer, because I am not an expert in Bioinformatics. I did find an interesting web page, though, that seems to be useful:
This page has a brief description:
SOURCE is a unification tool which dynamically collects and compiles data from many scientific databases, and thereby attempts to encapsulate the genetics and molecular biology of genes from the genomes of Homo sapiens, Mus musculus, Rattus norvegicus into easy to navigate GeneReports.
The mission of SOURCE is to provide a unique scientific resource that pools publicly available data commonly sought after for any clone, GenBank accession number, or gene. SOURCE is specifically designed to facilitate the analysis of large sets of data that biologists can now produce using genome-scale experimental approaches.
A more detailed description of SOURCE appears in
- SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, Alizadeh AA. Nucleic Acids Res 2003: 31(1); 219-23. [Medline] [Abstract] [Full text] [PDF]
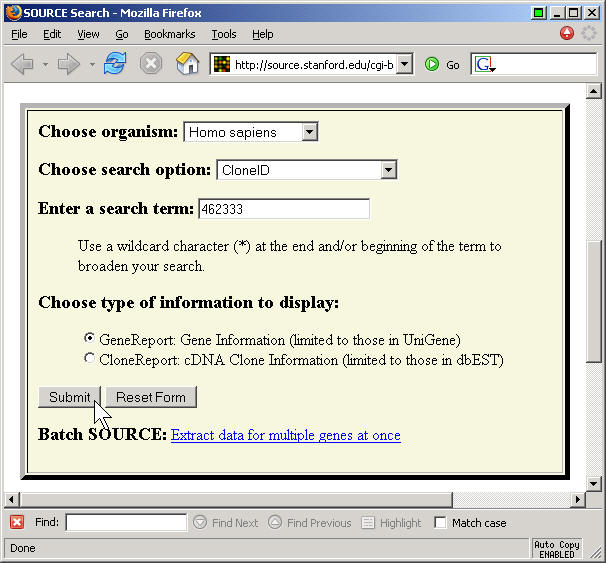
Taking the top gene on the list (462333), I ran a search:

and got a lengthy description of this gene. It is WD repeat domain 51A, or WDR51A. It has UniGene Cluster name Hs.476306 and also goes by the name NM_015426 (I think this is the GenBank name, but I am not 100% sure).
The information also listed the normalized expression distribution for this gene:
- pluripotent cell line derived from blastocyst inner cell mass
- mammary_gland
- thymus
- human embryonic stem cells
- thyroid
- placenta
- bone_marrow
- ovary
- stomach
- colon
Notice that ovary is on this list, but testis is not.
When you try the next gene on the list (2476256), you get the following message:
No Gene Information was found matching your cloneid query 2476256.
There is also a batch submission form which allows you to choose certain fields for extraction. This page is a bit fussy, because you have to put each CloneID on a separate line with no leading blanks.
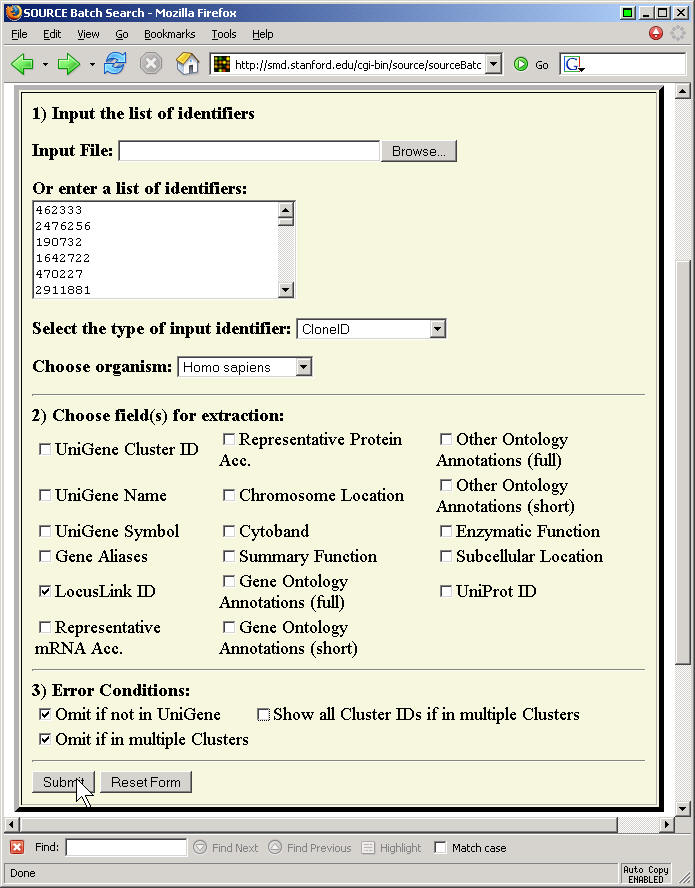
Here's a portion of the text file that this software produced
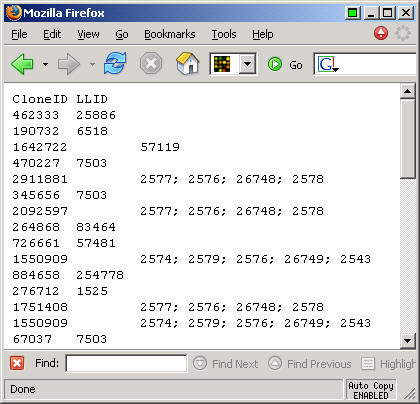
This looks like a tab delimited file, though the semicolons separate multiple values for the LocusLink IDs.