When you are importing a file from one system to another, lots of little things can trip you up. Here's an example, and it shows a very subtle problem.
I had an Excel file of infection events. The file looked roughly like this
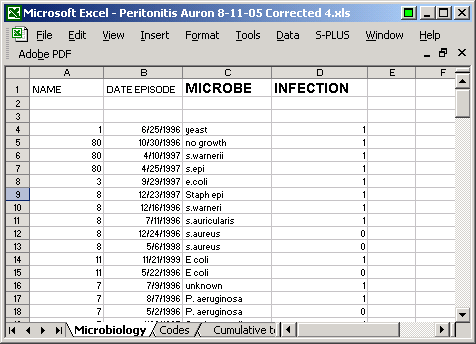
though I did change the dates to preserve confidentiality. To import this data into R, it is easiest to save the Excel file in a Comma Separated Value (.csv) format. When you do this, you get a few stray commas in the file.
NAME,DATE EPISODE,MICROBE,INFECTION
1,6/25/1996,yeast,1
80,10/30/1996,no growth,1
80,4/10/1997,s.warnerii,1
80,4/25/1997,s.epi,1
3,9/29/1997,e.coli,1
8,12/23/1997,Staph epi,1
8,12/16/1996,s.warneri,1
8,7/11/1996,s.auricularis,1,
8,12/24/1996,s.aureus,0,
8,5/6/1998,s.aureus,0,
11,11/21/1999,E coli,1,
11,5/22/1996,E coli,0,
7,7/9/1996,unknown,1,
7,8/7/1996,P. aeruginosa,1,
7,5/2/1996,P. aeruginosa,0,
Notice that partway down the file, an extra comma appears at the end of the line. There are a variety of reasons this might occur, but I ignored them because they did not seem to interfere with the importing of the data. I did clean up the extra two blank lines at the beginning, and imported the data in R using the read.csv() function. Here's what I got
NAME DATE.EPISODE MICROBE INFECTION
1 1 6/25/1996
yeast 1
2 80 10/30/1996 no growth
1
3 80 4/10/1997 s.warnerii
1
4 80 4/25/1997
s.epi 1
5 3 9/29/1997
e.coli 1
6 8 12/23/1997 Staph epi
1
7 8 12/16/1996 s.warneri
1
8 8 7/11/1996 s.auricularis
1
9 8 12/24/1996 s.aureus
0
10 8 5/6/1998
s.aureus 0
11 11 11/21/1999 E coli
1
12 11 5/22/1996 E
coli 0
13 NA
NA
14 7 7/9/1996
unknown 1
15 7 8/7/1996 P. aeruginosa
1
16 7 5/2/1996 P. aeruginosa
0
What caused the NA's to appear in row 13? Well, I first thought it was the stray commas, but they were dozens of them and the NA row appeared only once. So I decided to get rid of the stray commas by replacing any comma that appeared at the end of a line with nothing (using the regular expression ,$). That removed all but one of the stray commas. Then I realized that this particular comma was followed by a blank space, and R took that to represent a second new observation appearing on the same line as the previous observation. It only had a blank for NAME, which was converted to NA, and then since there were no more commas, it filled the remaining variables with blanks (if they were strings) or NAs if they were numbers.
So a stray comma caused no problems, but a comma followed by a blank (which is pretty hard to notice when you are skimming over the file) does cause problems.
This is not intended as a criticism of R or of the Comma Separated Value format, but just a reminder that converting data from one format to another can have unintended consequences. Always do a quality check on your data after you import it to make sure that nothing funny is happening.